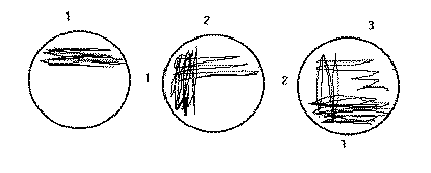VIRTUAL MICROBIOLOGY LAB
GUIDE
Table of Contents:
General Introduction
Objectives
Expectations
Introduction to Diagnostic
Laboratory Exercise
Bacterial
Morphology
Diagnostic
Microbiology
The
Gram Stain
Streaking
a Culture Plate for
Isolated Colonies
Specific Instructions
Part 1 - Gram-positive cocci
Blood agar
plates and hemolysis
Catalase test
PYR test
CAMP test
Bacitracin test
Binax Strep A (point-of-care) test
Optochin test
Coagulase test
Part
2 - Gram-negative rods
MacConkey Agar Plate
Citrate Slant
GENERAL
INTRODUCTION
I. Objectives
of the laboratory exercises
A. Provide familiarity with procedures
used for culturing (growing)
and identifying microorganisms of medical importance.
B. Aid in proficiency in submitting specimens for
the identification
of infectious disease in your future patients.
C. Provide clinical cases for diagnosis of
infectious disease and its
management.
These goals will be accomplished in the virtual microbiology
lab exercise
dealing with respiratory infection by identification of microorganisms
in
samples corresponding to case histories from patients. You will be
asked to
answer questions concerning the case histories and the general
principles of
diagnosis and of the etiological agents of respiratory diseases. For
those who
choose, practical experience can be obtained in streaking plates and
making Gram stains
in the limited wet lab.
II. What
is expected of you
Your completion of the virtual lab giving you practical
(decision making) and
theoretical experience is important because:
1. You will be tested for your
proficiency at reading and interpreting the results
(note that the virtual exercise is sufficient, since your exam is
electronic as well).
2. You will be tested on the theory
behind the etiological agents and their identification.
3. You are expected to record the results of tests, identify the
bacteria in the cultures, and arrive at diagnoses of the diseases.
4. You must submit your identifications and diagnoses along with
completing an online (open note, open book) homework assignment.
INTRODUCTION TO DIAGNOSTIC
LABORATORY EXERCISES
I.
Bacterial morphology.
Bacteria are 100-1000 times smaller than most mammalian cells;
they range from 0.4 to 3 microns (10-3 mm) in
diameter and
several microns in length. To examine them you will need a light
microscope with an oil immersion objective (100X).
Microorganisms differ widely in shape and size. The majority of
bacteria are either spheres (cocci) or rods
(bacilli). A few
occur as curved rods (vibrios) or in more complex shapes. Specific
types of bacteria may also vary in size and grouping
(single,
clumps,
pairs,
chains,
etc.). The shape and arrangement of the cells with their staining
properties are used for
classifying and preliminarily identifying clinical isolates. In
response to a hostile environment, some bacteria adopt a dormant
state by generating a spore, easily visualized with the light
microscope.
II.
Diagnostic Microbiology.
To identify the causative agent in infections, specimens are
obtained, and each organism is isolated and identified.
Microscopic examination of the specimen or the organisms is a first
step. This can be an unstained ("wet mount") specimen
or fixed specimen on a glass slide stained to visualize the
microorganisms and other cellular elements.
The most commonly used stain is the Gram stain, although other special
stains (e.g., acid-fast stain) can be used to tentatively identify
certain
organisms.
To grow and isolate microorganisms, the specimen is spread
("streaked") onto agar media containing nutrients to yield
colonies representative of each of the bacteria. Potentially important
organisms are
further tested for identification. A variety of methods is used -
culture media which select for growth of groups of organisms
(selective media), media containing indicators which
cause different organisms to appear differently (differential
media),
biochemical tests, phage typing, antibody typing, and many others.
III.
The Gram Stain.
A. Introduction.
The Gram
stain is one of the most valuable and most generally
used.
The Gram stain divides bacteria into two groups, the gram-positive
organisms, which stain dark purple to black, and the gram-negative
organisms, which take on the color of the counterstain, usually red.
Bacteria are stained with crystal violet
followed by Gram's iodine. These two solutions form a complex which, in
gram-positive bacteria, is not washed away with
ethanol; gram-negative bacteria rinse clear. To visualize the clear
gram-negative bacteria, they are counterstained with a
contrasting color. Red stains (e.g., safranin) are usually used.
The ability of gram-positive
bacteria to retain the crystal violet-iodine complex following
treatment with ethanol varies with
the age and species of bacteria and, to a lesser extent, the
environment from which they were obtained.
B. Procedure for Gram staining a
specimen.
UF Students note - You need to understand the steps of the Gram stain
and what happens at each step fr gram-positive and gram-negative
bacteria, but you do not need to memorize the times.
1. Using a sterile loop, transfer a loopful of tap water to a
clean glass slide. Touch a loop to the desired colony,
and mix the bacteria in the water on the slide. A VERY SMALL amount of
bacteria will suffice.
2. Allow the specimens to dry on the slide at room temperature. Do
not heat the slide to speed drying because this can
distort the cellular morphology or staining properties of the
organism.
3. After the specimen has dried, heat-fix the slide. Gently
heat the slide by passing quickly through the flame, specimen side
up, 3-4 times. It should be warm but not hot to the touch.
4. Stain the bacterial smears by Gram's method as follows:
Flood the slides sequentially with solutions a-d for the indicated
times.
(a) Crystal Violet--------------------------------1
minute
Wash gently in tap water for 2-3 seconds.
(b) Gram's Iodine (I2-KI)-------------------------1
minute
Wash gently in tap water, shake off excess water.
(c) 95% alcohol-----------------------------------10
seconds
Do not over-decolorize the specimen with alcohol. If you're
going to screw up the Gram stain, this is the step!
Wash gently in tap water, shake off excess water.
(d) Safranin (counterstain)-----------------------20
seconds
Wash in tap water and blot dry.
5. Examine with oil immersion optics (not at lower
power). Move the condenser almost all the way up to touching
the slide.
Do not let the high/dry (40X) lens get into the oil. It will be very
difficult to clean.
6. Gram-positive
organisms will be purple/blue. Gram-negative organisms
will be pink to red.
IV.
Streaking a plate for isolation of colonies.
A. Introduction.
The single
most important step in analyzing a specimen containing
bacteria is to obtain
isolated colonies of bacteria
that arise from single cells. Attempts to identify bacteria in a
clinical sample cannot be done unless isolated colonies are
used.
To obtain well-isolated
colonies, it is essential to disperse the inoculum (sample) on the
surface of an enriched agar plate so
that individual bacteria are well separated from each other. Ideally,
each of the bacterial species present will produce a
distinct colony type. The appropriate technique will be demonstrated by
one of the instructors in each laboratory.
B. Procedure.
1. With the loop, spread the inoculum back and forth across the upper
1/4 of the plate, keeping the lines of inoculation very
close together (area 1 in this figure). Isolated
colonies are not expected in this area. Do not use strong pressure,
which will
break the surface of the agar. Use the end of the loop, not its side
when streaking.
Dispose of the loop in the biohazard bucket on the bench.
2. Turn plate approximately 90 degrees. Streak the plate across about
1/4 of the plate. (See area 2 of the figure.)
Dispose of the loop.
3. Turn the plate 90 degrees again, using the loop streak into
the second area only a couple of times and then zig-zag across
the remaining open area of the plate - being sure not to cross into
areas 1 or 2 as this will put too many bacteria into this
area that should hopefully contain isolated colonies. Stab
the first streak area a couple of times to accentuate hemolysis.
4. Label plates and incubate inverted at 37 C.
Single
colonies should appear
in area 3.
Note: in drawings, lines should be closest together in Sec. 1
and progressively further apart in succeeding sections.
SPECIFIC
INSTRUCTIONS
Part 1
Identification of Gram-positive Cocci
I. Introduction.
The gram-positive cocci include organisms that are round and
that usually occur in
chains or pairs (streptococci) and
those that occur in clusters or bunches (staphylococci). Infections
by pathogenic gram-positive cocci are responsible for
many bacterial diseases, ranging from superficial skin lesions to
severe life-threatening infections. Other members of the
group are fairly regular inhabitants of skin and mucous membranes, the
so-called "normal flora."
Blood agar plates.
The primary isolation from infectious material is usually made on sheep
blood agar, a rich medium that supports the growth
of many types of microorganisms. The appearance of colonies and red
blood cell lysis are important diagnostic features.
The most common streptococci and staphylococci can be divided into
groups on the basis of their reactions on blood agar
(examples are shown at
labimage/imagky.html
on the MMID home page):
Alpha hemolytic -
partial lysis of red blood cells, producing a greenish
discoloration. The two most important groups
are Streptococcus pneumoniae (pneumococcus), a
frequent cause of lobar pneumonia and several other serious infectin, and the viridans group of
streptococci, normal inhabitants of the oropharynx that may cause
disease (e.g., endocarditis) when they invade the vascular
system. You will ALWAYS have viridans streps in a throat swab.
Beta hemolytic -
complete lysis of red blood cells and clearing of the
medium around the colony. Common pathogens
which produce this reaction are Groups A, B, C and some D streptococci,
as well as Staphylococcus aureus, the most
common pathogenic staphylococcus. For this lab, we are focusing on Group A strep (Streptococcus pyogenes) and Group B strep (S. agalactiae).
Gamma hemolytic
- no apparent change in the medium (non-hemolytic is
more descriptive). Staphylococcus epidermidis,
a normal skin inhabitant. (See the insert of this figure
to compare gamma and beta hemolysis.)
II. General Procedures.
The purpose of this experiment is to make observations of some
diagnostic features of the important streptococci and
staphylococci.
A. Differentiation of
streptococci from staphylococci.
Although microscopic examination of stained smears
presumptively permit distinction between these two groups of
organisms, a definitive classification can be made on the basis of the
presence or absence of the enzyme catalase. Staphylococci
contain this enzyme, streptococci do not.
Catalase
test. Place a drop of 3% hydrogen peroxide
on a clean microscope slide. Place a heavy loopful of cells from
isolated colonies into the liquid (you may have to pick up four to five
colonies if they are small). Immediate generation of
gas bubbles constitutes a positive test. Avoid the inclusion of blood
cells from blood agar plates as blood contains catalase.
Lack of bubbles is a negative test. (Picture of results.)
catalase
2H2O2
--------------------> 2H2O + O2
(bubbles)
B. Identification
of group A Streptococcus (Streptococcus
pyogenes).
1. PYR test.
The PYR test has replaced the bacitracin test as the
culture-based test for Group A streptococcus (S. pypogenes). It
detects the presence of the enzyme Pyrrolidonyl
Arylamidase. Pyrrolidonyl
Arylamidase cleaves β-naphthylamide from a L-naphthylamide-β-naphthylamide
substrate, and the released β-naphthylamide reacts with N, N-dimethylaminocinnamaldehyde present in a test strip
yielding a red/pink color. The test is done by placing some
colony material from a plate onto the test strip and looking for the
color change. (Picture of results) In contrast, other beta-hemolytic
streptococci, most notably Group B streptococcus (S. agalactiae), are
PYR-negative. The PYR test is done in conjunction with the
CAMP test, since it yields complementary results.
2. CAMP test.
The CAMP test detects the presence of the CAMP factor (an
acronym for the names of the discoverors), which is produced by some
gram-positive cocci. The CAMP factor potentiates hemolytic
activity produced by Staphylococcus
aureus. Group B strep is CAMP+, whereas Group A
strep is CAMP-. The assay is performed by streaking S. aureus down the
middle of a blood agar plate. The unknown beta-hemolytic
strep is cross-streaked across the staph, and the plate is incubated.
If the strep produces CAMP factor, an arrowhead-shaped zone
of heightened hemolysis will appear at the junction of the streaks,
(Picture of results) The CAMP test is done in conjunction with the PYR test.
3.
Bacitracin test.
The bacitracin test used to be the standard culture-based method for
confirming S.pyogenes
from other beta-hemolytic streptococci, such as Group B strep (S. agalactiae).
However, it has been replaced by the PYR and CAMP tests.
You should be familiar with the bacitracin test since it
could show up on standardized exams. Commercially available
paper disks saturated with a solution containing
Bacitracin will inhibit about
97% of all strains of Group A streptococci; other groups of
beta-hemolytic streptococci will not be affected. Streak a blood
agar plate with an isolated colony of beta-hemolytic streptococci
(you're not looking for isolated colonies now). After
inoculation, flame the provided forceps, and aseptically pick up a
bacitracin disk (B or A disk).
Place the disk on the plate and press gently onto the agar medium to
ensure firm contact with the agar. Observe the plates for inhibition of
growth(indicating sensitivity) after overnight incubation at 37.
For Streptococcus
pyogenes there will be a zone of inhibition of growth around
the A disk. Note that the hemolysin might diffuse in the agar
and make it look like the bacteria grew closer to the disk than
reality. Also note that over inoculating the plate will make it
difficult to interpret as well as under inoculating.
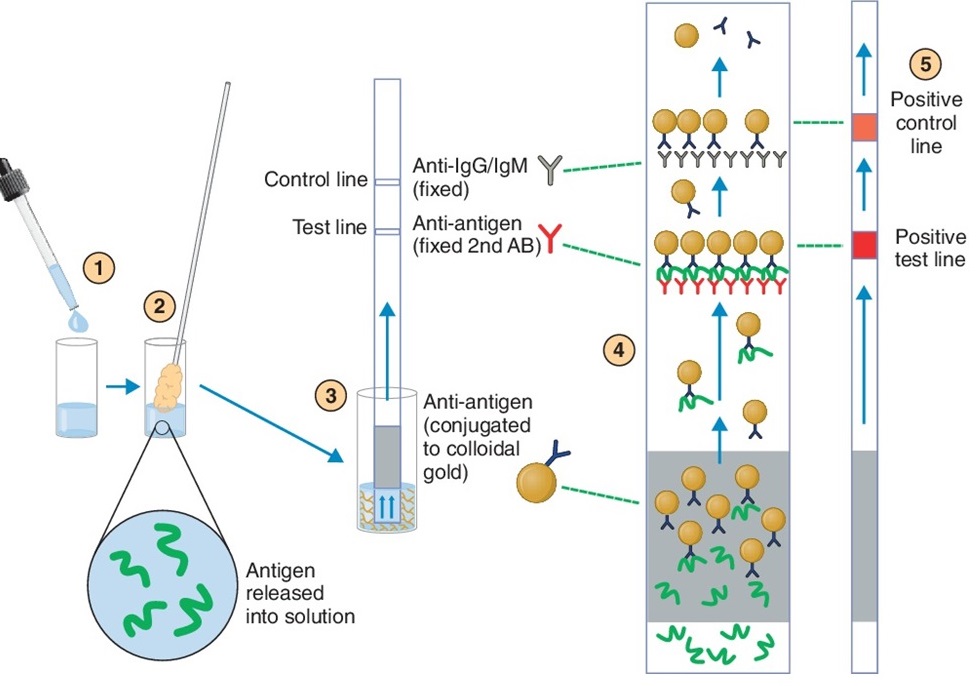
4. BINAXNOW™ STREP A
test. This is a point-of-care rapid test that is essentially the
same as the at-home COVID-19 tests that we have all become familiar with as a
result of the pandemic. The
test is detecting ANTIGEN using antibodies.
A swab is used to obtain a sample from the relevant site. For Group A streptococcus pharyngitis, it
would be the back of the throat. For
COVID-19, it is the nose. The material
in the swab is released with a buffer.
For this kit, the buffer is added to a well in the test card immediately
before the test is run. The swab is
mixed with the buffer in the card.
Here's how the kit works (see the
figure). The buffer contains gold
particles that are linked to the detection antibody, in this case anti-Group A
carbohydrate. If the sample has Group A
carbohydrate, it will bind to the gold particles via the anti-Group A
antibodies. The solution migrates up the
strip in the card via absorption. The
test line in the strip contains anti-Group A antibodies. If the sample has Group A antigen, it will
bind to the test line of anti-Group A antibody and capture the attached gold
particles (the Group A carbohydrate is polymeric and can bind to more than one
antibody). This will form a red line on
the test line. This is necessary, but
not sufficient, for a positive result.
Above the test line is a control
line. It has anti-IgG/IgM
antibodies. These antibodies will bind
to the anti-Group A:gold particles and cause a red line, even if there is no
Group A antigen in the sample. Note that
there will be plenty of anti-Group A:gold particles to enable this to happen,
if it is a positive sample. Failure of
the red line to appear in the control line indicates a failure of the
test. This is most important if a
negative result is obtained in the test line.
If the control does not turn positive, the negative test is
meaningless. Getting a positive test but
negative control is unlikely, but would still invalidate the test.
This test is very specific
because of the use of specific antibodies, and it is reasonably sensitive;
however, it is possible that there is an infection that is low enough to not
cause a positive result or that the sample was not adequate (poor
swabbing). Therefore, a negative test in
suspected Group A strept pharyngitis should be followed with a more sensitive
test such as culturing. This is true for
all point-of-care tests like this.
C. Differentiation of
pneumococci from other alpha hemolytic streptococci.
Optochin
test. Pneumococci (but not
other alpha-hemolytic streptococci) are inhibited by optochin.
Apply a disk of filter
paper containing optochin (O or P disk) to a heavily streaked plate
(see procedure for applying the bacitracin disk). If the
organism is a pneumococcus,
a
large zone free of bacterial growth will surround the paper disk
(indicating sensitivity) after overnight incubation at
37C. If the organism is another alpha-hemolytic streptococcus, there
will be no
zone of inhibition around the disk, or at most
a narrow one. Optochin is available commercially as "Taxos P" or
"Optochin disk."
D. Differentiation of Staphylococcus
aureus from non-pathogenic staphylococci.
Coagulase
test. The test which
distinguishes S. aureus from non-pathogenic
staphylococci is the coagulase test. Coagulase
is a secreted enzyme of Staphylococcus aureus
that causes plasma to clot (coagulate). The tube test is performed by
inoculating an isolated colony into 10% rabbit plasma. After incubation
at 37oC for 2-4 hours, examine the tube for the
presence of (picture).
If the reaction is negative, incubate overnight and re-examine the tube.
Part 2
Identification of
Gram-negative enteric rods
I. Introduction.
This part of the exercise will focus on the family Enterobacteriaceae,
which includes several genera of medical importance.
This is a large and diverse group, and the laboratory methodology for
their identification has evolved over many years. You
will work with Klebsiella pneumoniae and Escherichia
coli.
The choice of medium for the initial isolation from a clinical
specimen may depend on the specimen source. Usually
specimens are cultured initially both on blood agar and a number of
SELECTIVE media (e.g.,
MacConkey agar, which
excludes the growth of gram-positive organisms because it contains bile
salts). Media such as MacConkey also permit
an assessment of the ability of the organisms to ferment lactose
(DIFFERENTIAL media), which provides one of the key
branch points in the diagnostic scheme.
These days systems are used which enable the simultaneous
inoculation of many media and the evaluation of numerous
biochemical characteristics. In this laboratory exercise, we will use
the more classical (old fashioned) techniques because
they form the foundation for all metabolic identification systems.
II. General Procedures
A. MacConkey
agar plate. Streak samples
for isolated colonies on MacConkey agar using same procedure as for
Part 1.
MacConkey agar is an example of a selective medium; it permits growth
of gram-negative enterics but inhibits the growth of
gram-positive bacteria.
After 24 hours incubation, examine the plates to distinguish
the two different colonial types. The MacConkey medium
provides evidence as to whether each organism ferments lactose.
Colonies which ferment lactose are red. This reaction is
due to the action of acids produced by fermentation of lactose on the
bile salts and the subsequent absorption of neutral red,
a pH indicator, from the medium. Colonies of non-fermenters of lactose
appear colorless.
Example
of results of a mixture of
lactose-positive and negative bacteria on a MacConkey agar
plate.
E. coli and K. pneumoniae
are usually lactose-positive. To make life interesting, you will also
be given a lactose-negative mutant E.
coli as part of this exercise.
Also note that the E. coli and Klebsiella
pneumoniae in your mixtures are vastly under represented.
You will have a
difficult time finding them on your blooad agar plates since they will
be outnumbered by the staphs and streps.
However, on the MacConkey plate, on which the streps and staphs don't
grow, the gram-negative rods come forth
in their true colors.
B. Citrate
slant.
Some bacteria can use citrate as a source of carbon, while
others cannot.
Streak the surface of the citrate tube with an
isolated colony from the MacConkey plate. Incubate the tube with the
lid loose. Utilization of citrate as a carbon source
results in a color change from green to blue. The indicator is
bromthymol blue, which turns from
green
to blue at low pH.
Read the test at 24 hour incubation at 37C.
K. pneumoniae is citrate-positive, while E.
coli is usually citrate-negative. Both of your E.
coli strains are typical for
citrate utilization. In real life, as opposed to a contrived
Virtual Lab, differentiating E. coli and
Klebsiella involves an extensive battery of
metabolic tests which is used to differentiate the gram-negative enteric rods..
OK, I've read through this
stuff. I'm an expert. I'm ready to solve some
cases! (Click on the cases
in the side bar.)